Auto-Annotation
Try It Yourself (4 steps)
Click here to pull in the data for this tutorial. The sequence pCMV-eGFP will be added to a project called Training and opened for you.
We are going to use Benchling’s auto-annotation feature to add annotations to this sequence.
-
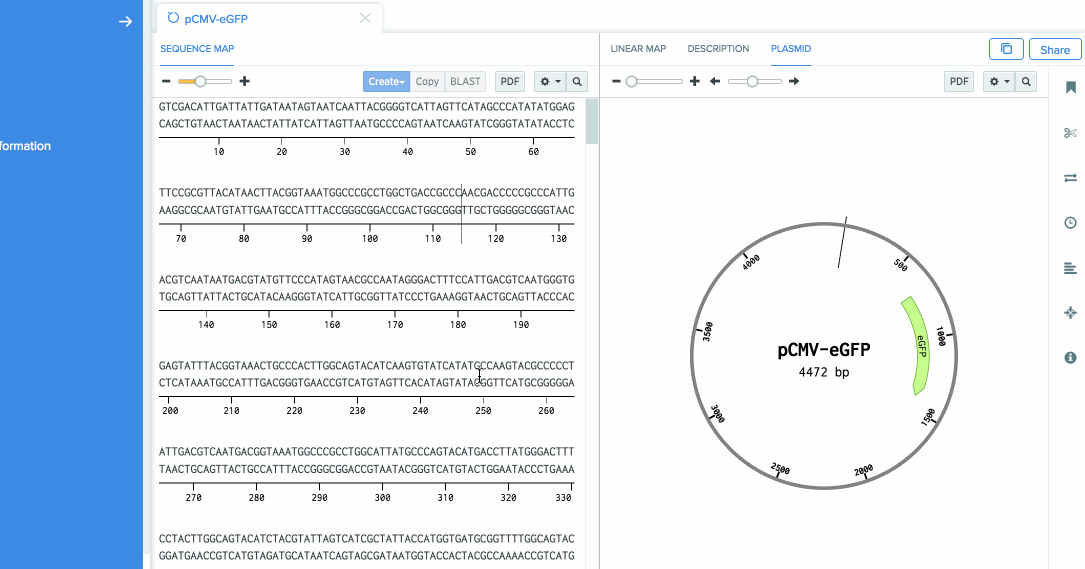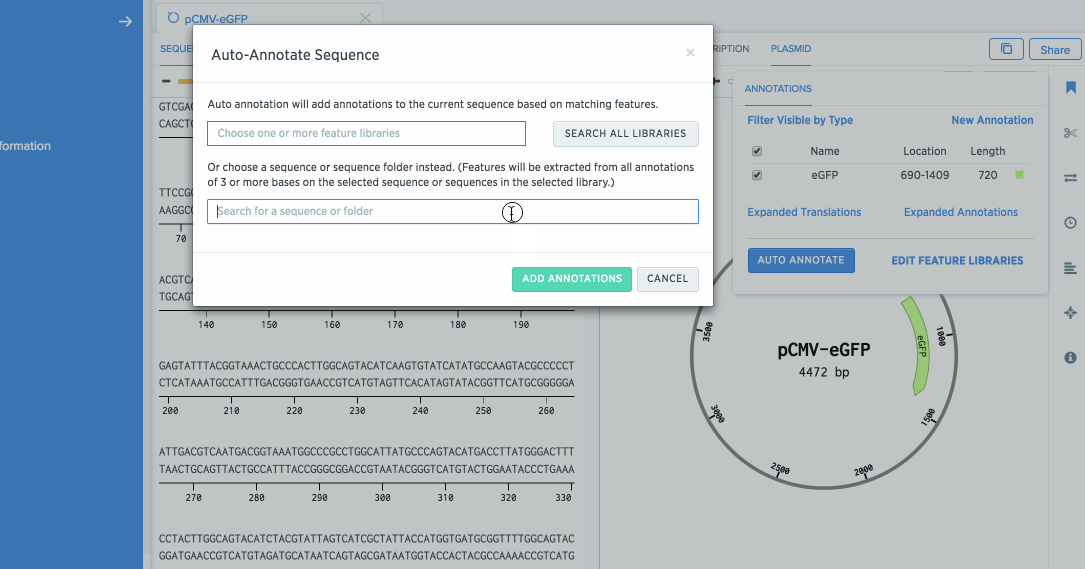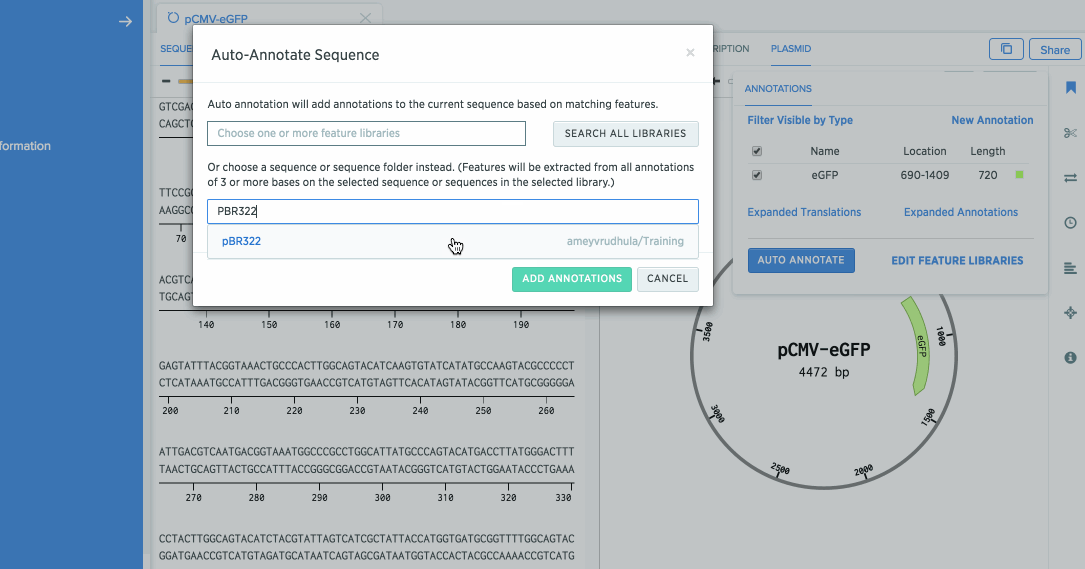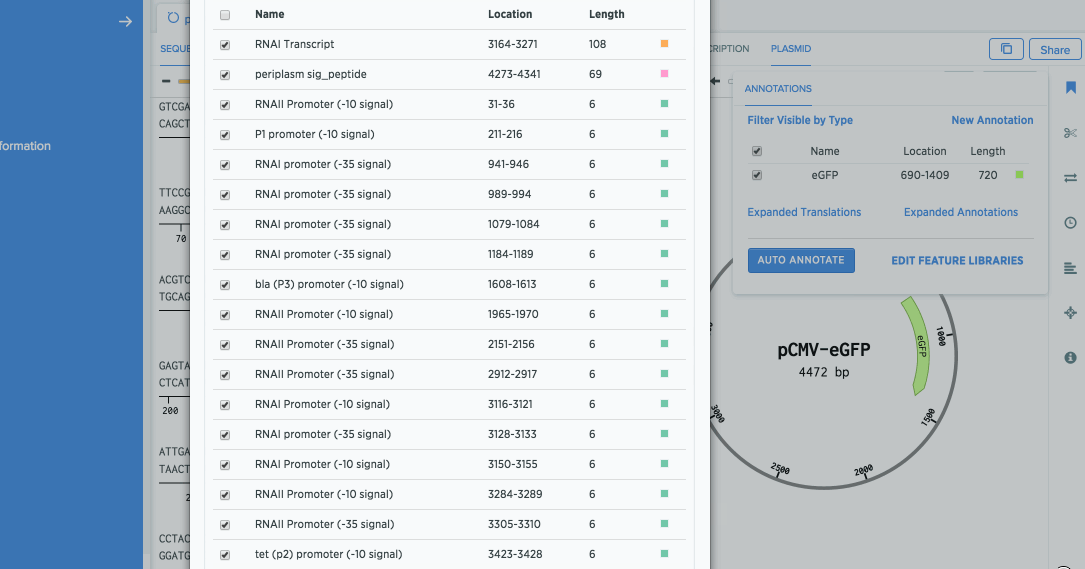
Open the annotation panel, and click on Auto Annotate.
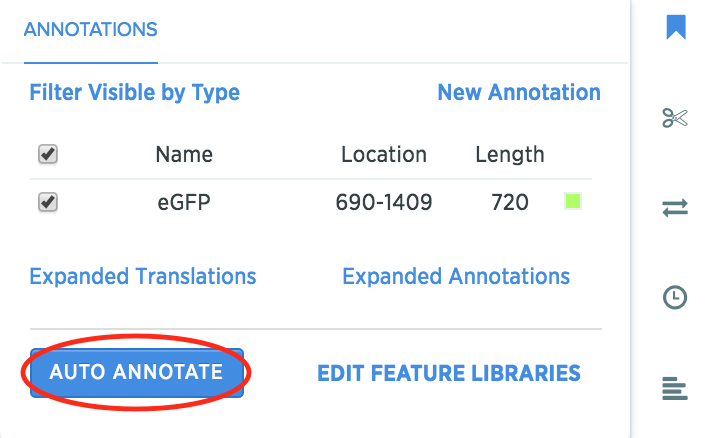
-
Click on Search All Libraries to search over all of your feature libraries. Benchling provides you with some default feature libraries. You can view these by clicking on your username at the top, and clicking Feature Libraries.
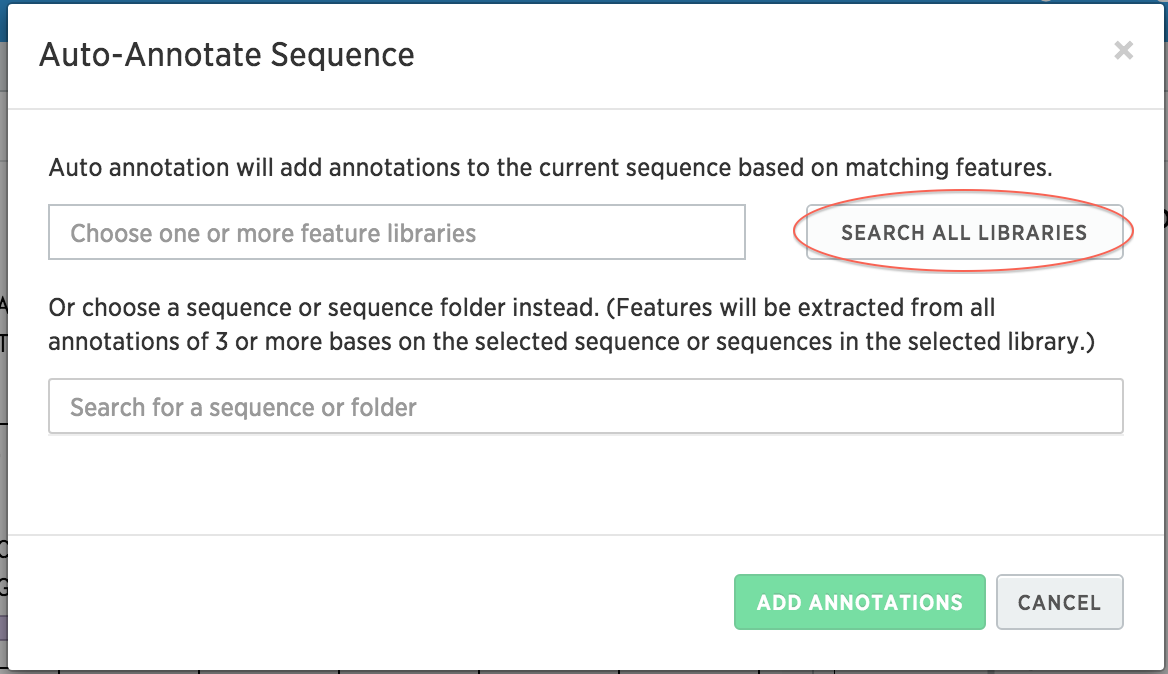
-
Select all the annotations, and click Add Annotations. You should now see several annotations added to your sequence.
-
Right click on the eGFP annotation, and click Add To Feature Library. This will add the eGFP annotation to one of your feature libraries, so you can use it for auto-annotation.
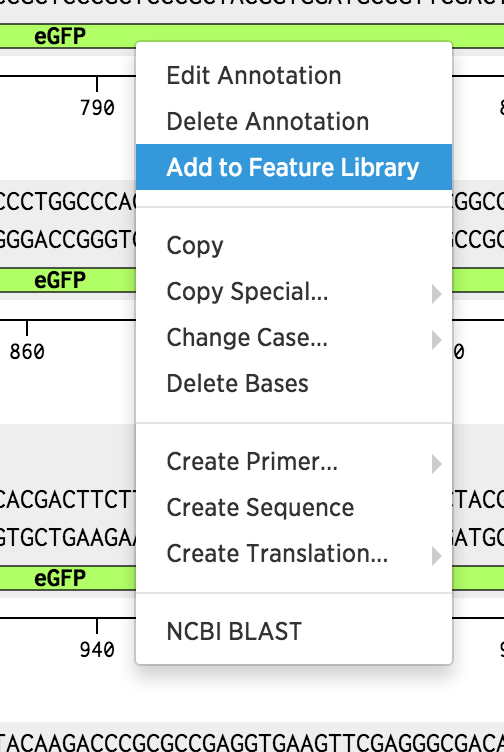
Pro Tip: You can use annotations from an existing sequence. Benchling will search for annotations in the sequence you enter and mark them in the sequence you are annotating.