Consensus Alignment
Try It Yourself (4 steps)
Click here to pull in the data for this tutorial. A folder called HBB will be added to your Training project. This folder contains four variants of the Human Myoglobin, Beta gene. We are going to do a consensus alignment on these.
We’ve expanded the folder for you already. (Clicking the Open in new tab button on the file browser expands a folder.)
-
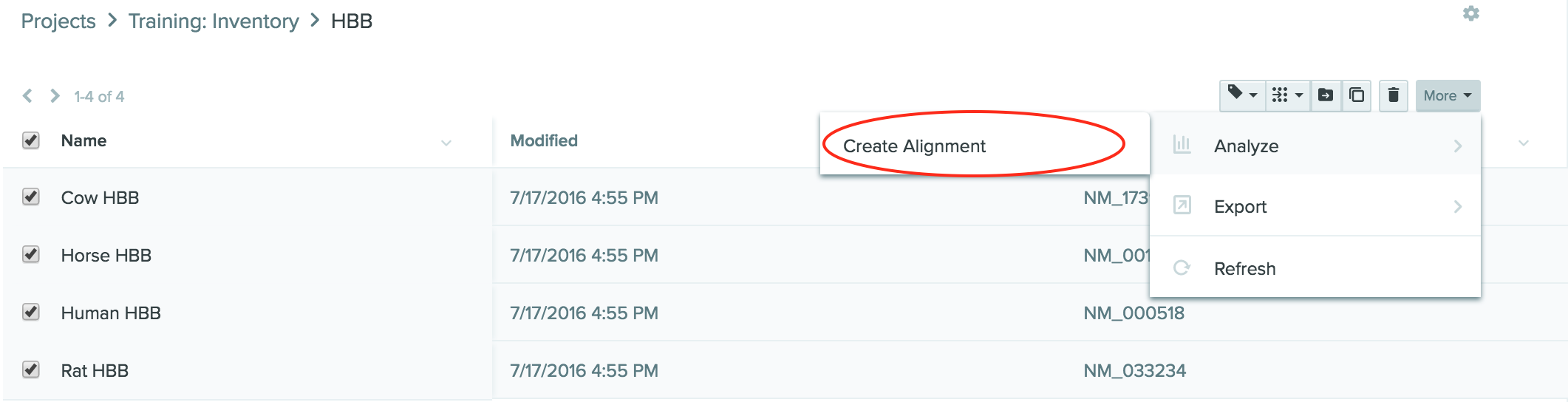
Select all the sequences, click the Create Alignment button on the top right, and save in the Training folder. You can also go to Create and select Create Alignment from the dropdown.
-
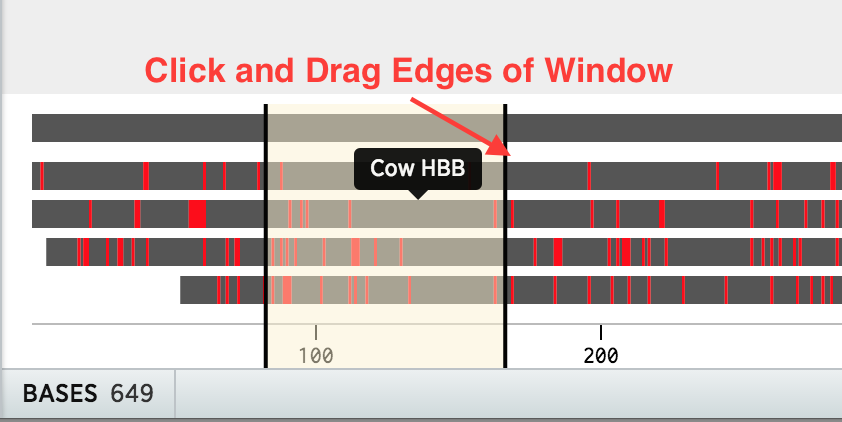
Once the alignment is complete, click and drag the bottom summary (grey bars) to view different regions. Zoom in by decreasing the width of the viewing window.
-
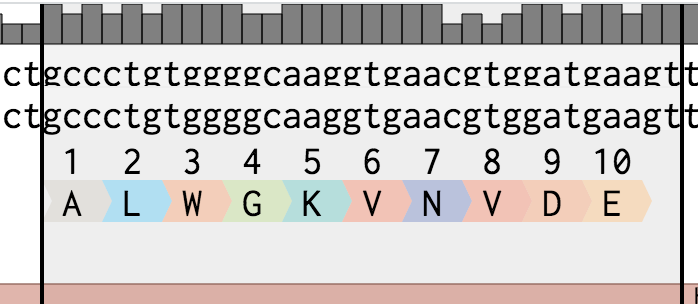
To create a translation, select a region of a sequence, right click, choose Create Translation, and click Forward.
-
Misalignments are shown in red in the summary. You can also add a comment by selecting bases and clicking Add Comment at the top of the tab.
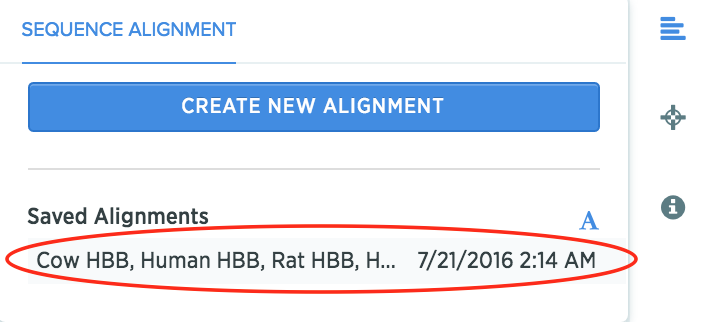
Pro Tip: View the consensus alignment again by going to the inventory file that was created, and clicking on the *Alignment *icon. Then click on the saved alignment.