Cloning by Copy and Paste!
Try It Yourself (3 steps)
See the Digest and Ligate to learn about using the Assembly Wizard to automate cloning.
Click here to pull in the data for this tutorial. The sequences pDusk and pDsRed2 will be added to a project called Training and opened for you.
We are going to clone the common fluorescent protein DsRed2-Express into the backbone pDusk. pDsRed2 has the protein flanked by unique cut sites.
-
Copy pDusk into a new sequence from the History panel (clock icon at the right). Name the new plasmid “pDusk-DsRed2” and save it into the Training project. This will become the assembled plasmid.
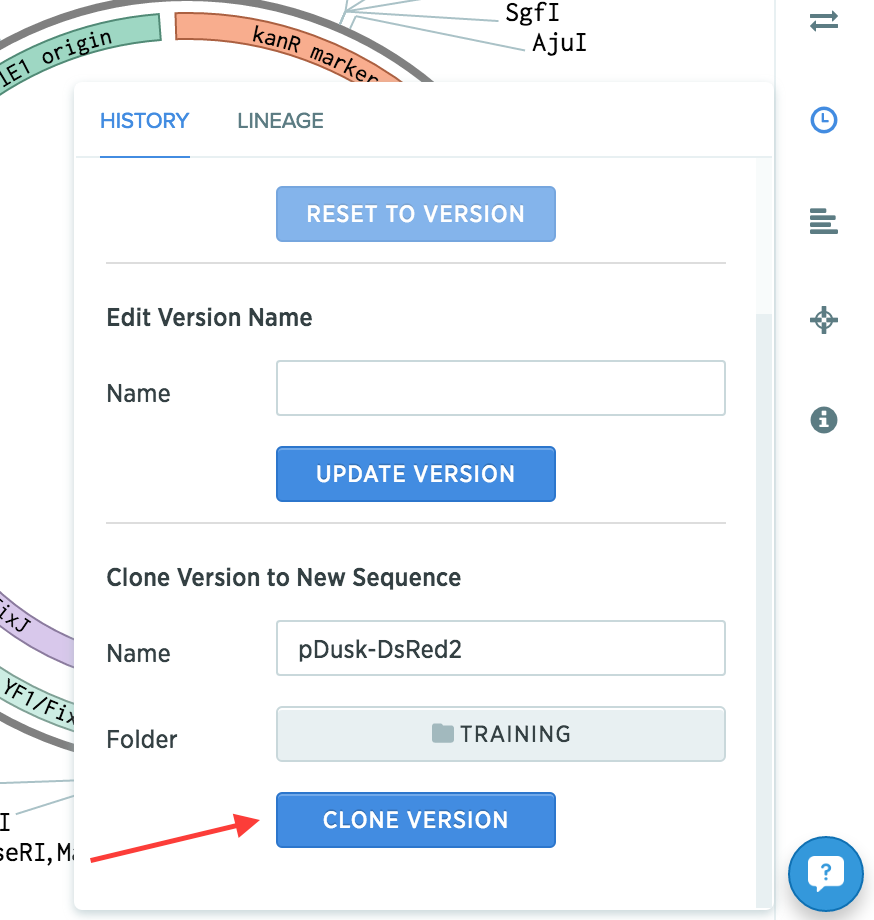
-
We’ll use BamHI and NotI for cloning. (For more information on how to find which enzymes to use, see this tutorial.)
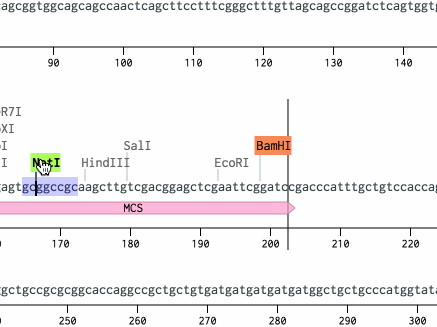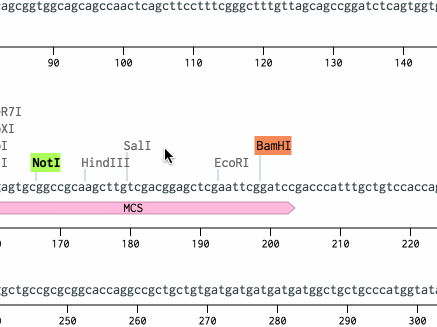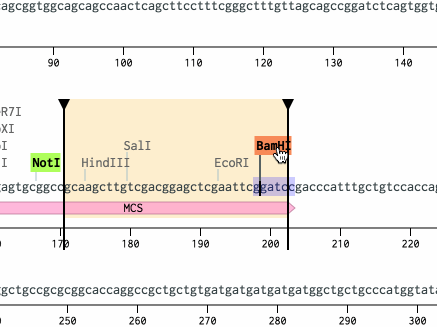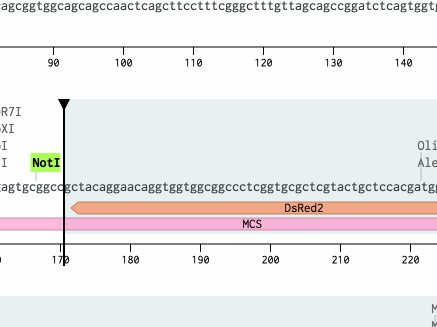
Open pDsRed2 from the workspace at the bottom left. Scroll down to the BamHI site at bp 265 and click it. Then, scroll down to the NotI site at bp 968, hold Shift, and click it. Then, hit Cmd+C (or Ctrl+C on a PC) to copy the region.
The BamHI and NotI cut sites should get highlighted. Benchling will helpfully highlight all cut sites with compatible sticky ends. (For other ways to turn this on, see this tutorial.)
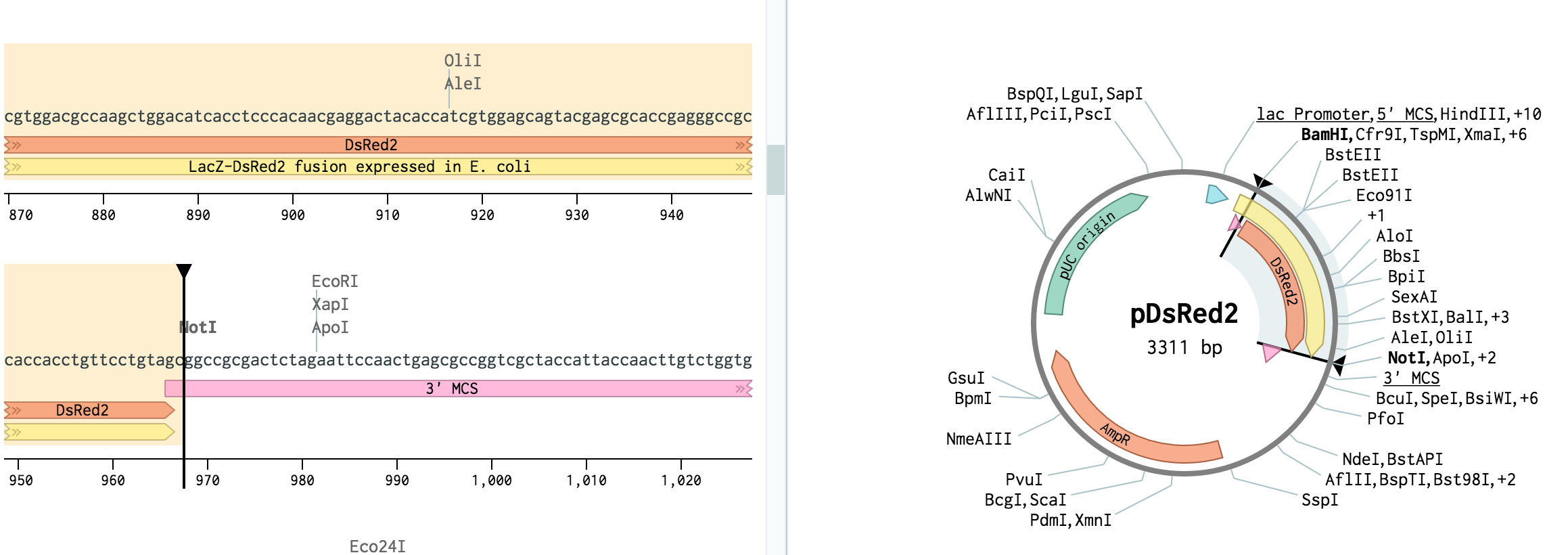
-
Reopen the pDusk-DsRed2 plasmid. Scroll down to bp 167 and shift-select the NotI and then BamHI sites. Hit Cmd+V (or Ctrl+V on a PC) to paste the region.
Notice that because you used cut sites to copy and paste, Benchling automatically reversed the fragment and handled ligating sticky ends. Annotations and translations from pDsRed2 were also brought along.